
A large-scale population metagenomic study has shed new light on the spatial heterogeneity of viral communities across the gastrointestinal tracts of ruminants, which are closely linked to human history.
The study, led by Prof. TAN Zhiliang from the Institute of Subtropical Agriculture of the Chinese Academy of Sciences, demonstrates that the gastrointestinal tract region, rather than ruminant species, is the primary factor that distinguishes viral communities.
The findings were published in Journal of Advanced Research on January 6.
In this study, the researchers constructed the Ruminant Gastrointestinal Virome Catalog (RGVC) by integrating 373 metagenomic samples from 10 gastrointestinal regions across seven ruminant species. By combining short- and long-read sequencing, they identified nearly 44,000 viral operational taxonomic units (vOTUs), 89.3% of which were previously unknown, profoundly expanding our map of the ruminant virosphere.
The analysis revealed a defining feature of these communities: pronounced spatial heterogeneity. Viral composition is primarily shaped by physiological gastrointestinal regions, such as the stomach, small intestine, and large intestine, rather than by host animal species. This regional patterning is closely linked to the putative microbial hosts. Bioinformatic analysis revealed over 10,000 high-confidence virus-host pairs between more than 4,600 prokaryotic hosts and nearly 6,000 viruses. A strong correlation was found between the abundance of viruses and their corresponding hosts, indicating that differences in the local prokaryotic community are the key drivers of viral distribution.
Further investigation showed that nearly half of the viruses employ a lysogenic lifestyle, integrating their DNA into the host genome for long-term persistence, with the dominance of this strategy varying systematically along the digestive tract.
In addition, many of the viruses were found to carry auxiliary metabolic genes (AMGs) associated with key metabolic processes, including carbohydrate and energy metabolism. The distribution of these AMGs differs across gastrointestinal regions, corresponding to their localized metabolic roles. Further studies confirmed the viral origin and functional activity of these key AMGs through genomic analysis and 3D protein modeling.
"As the first systematic exploration of ruminant gastrointestinal viruses through the lens of spatial ecology," said Prof. WANG Min, co-corresponding author of the study. "This work deciphers their distribution, survival strategies, and functional impacts, and enabling targeted modulation of virus-host interactions in specific gastrointestinal compartments."
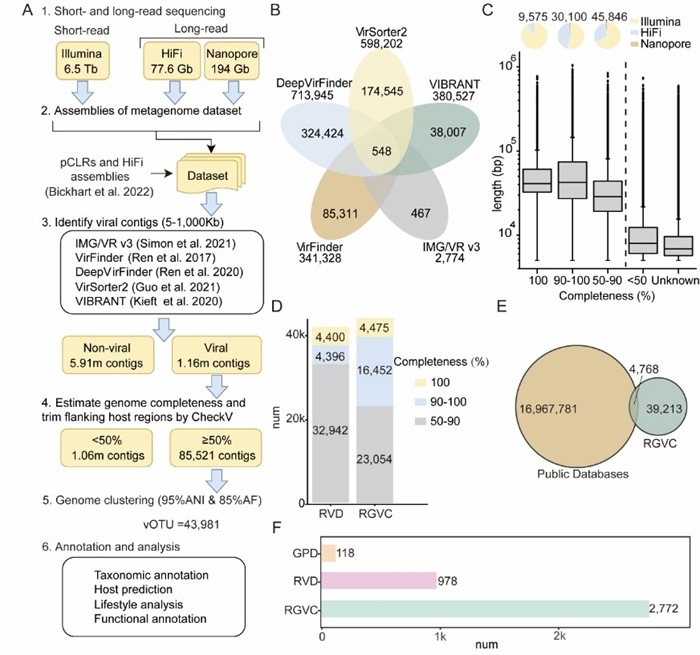
Construction and feature overview of the RGVC (Image by ZHANG Shizhe)
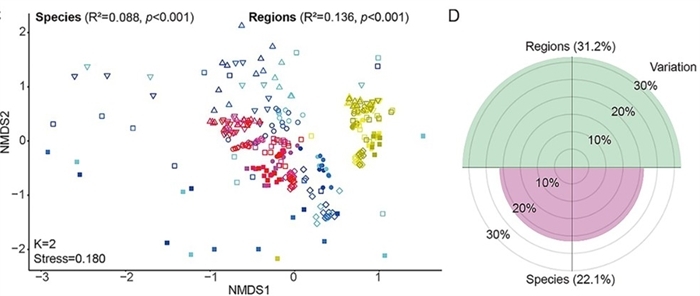
Viral diversity and proportion of variation explained by gastrointestinal region and ruminant species (Image by ZHANG Shizhe)
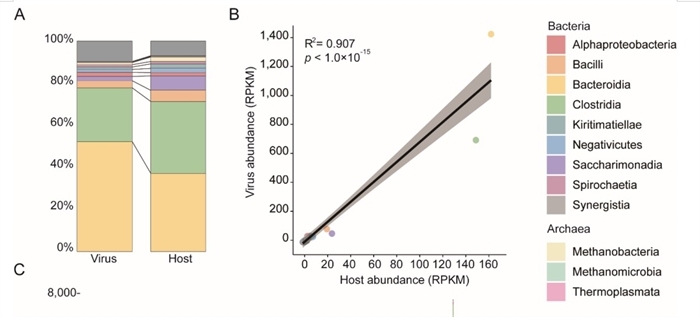
Dynamic correlation between viruses and their putative hosts (Image by ZHANG Shizhe)
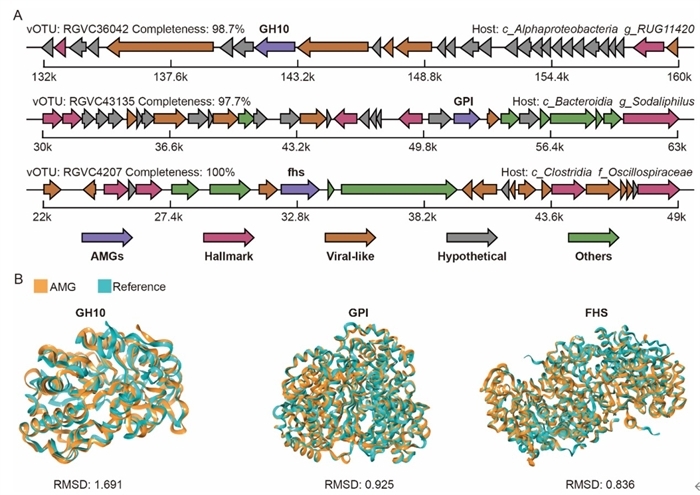
Genomic context and protein structure of selected virus-encoded auxiliary metabolic genes (Image by ZHANG Shizhe)

86-10-68597521 (day)
86-10-68597289 (night)

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

